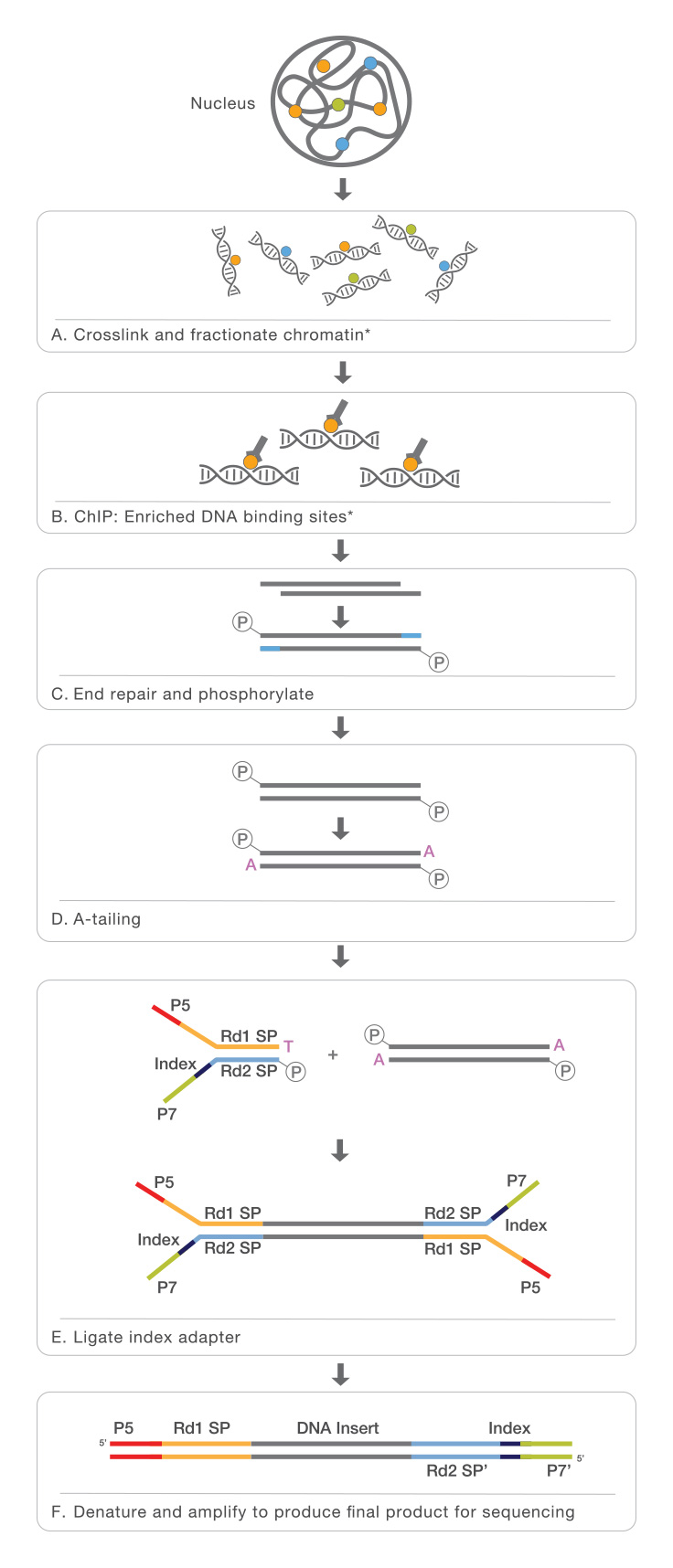
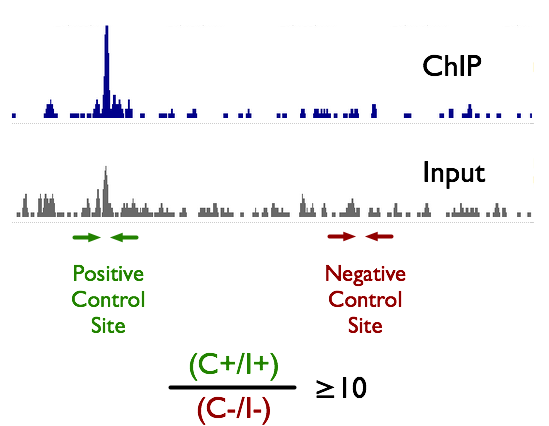
Improved mapping with ChIPSeq deconvolution. (A) Summary of ChIP-Seq... | Download Scientific Diagram

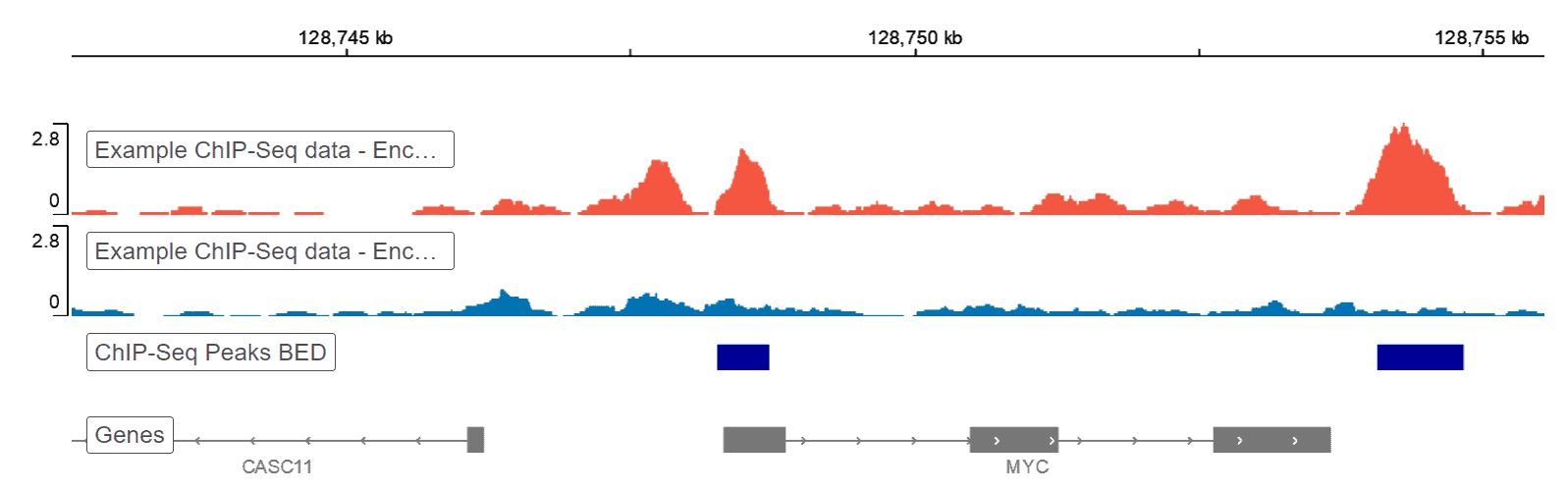
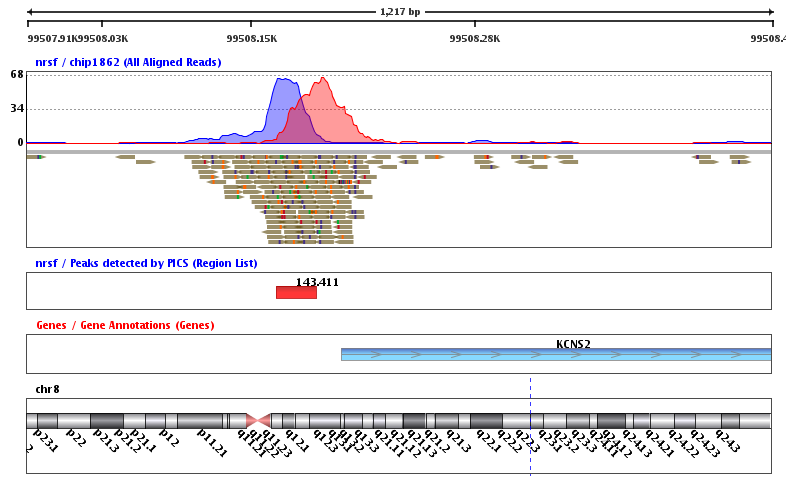
Example genome browser screen-shot. ChIP-seq read coverage of H3K27me3... | Download Scientific Diagram

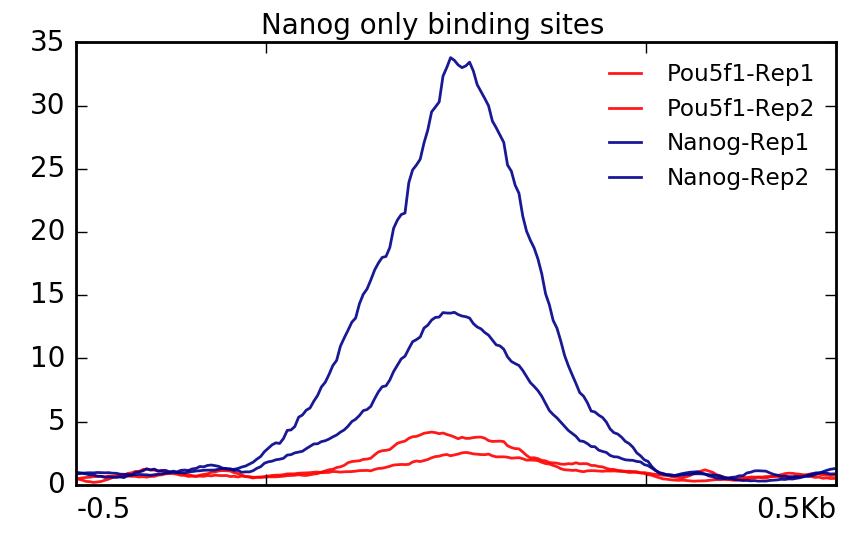
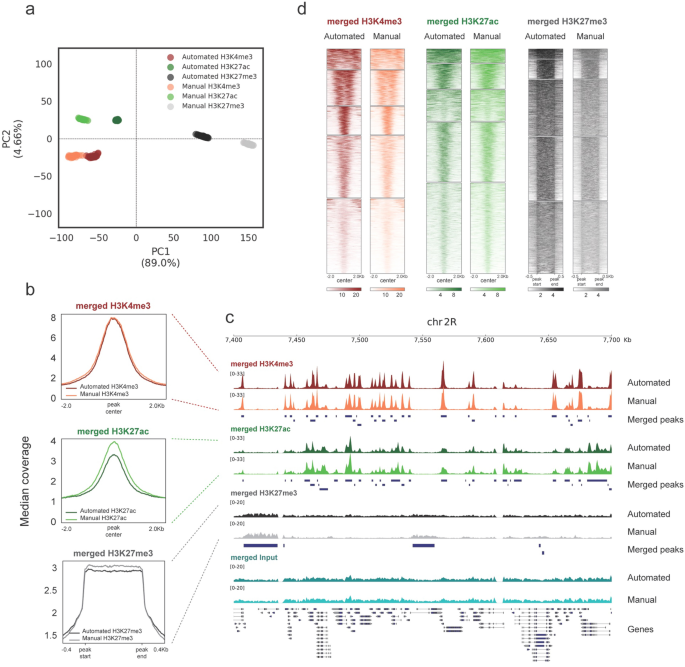
ChIP-Seq deconvolution maps are highly reproducible. (A) Comparison of... | Download Scientific Diagram

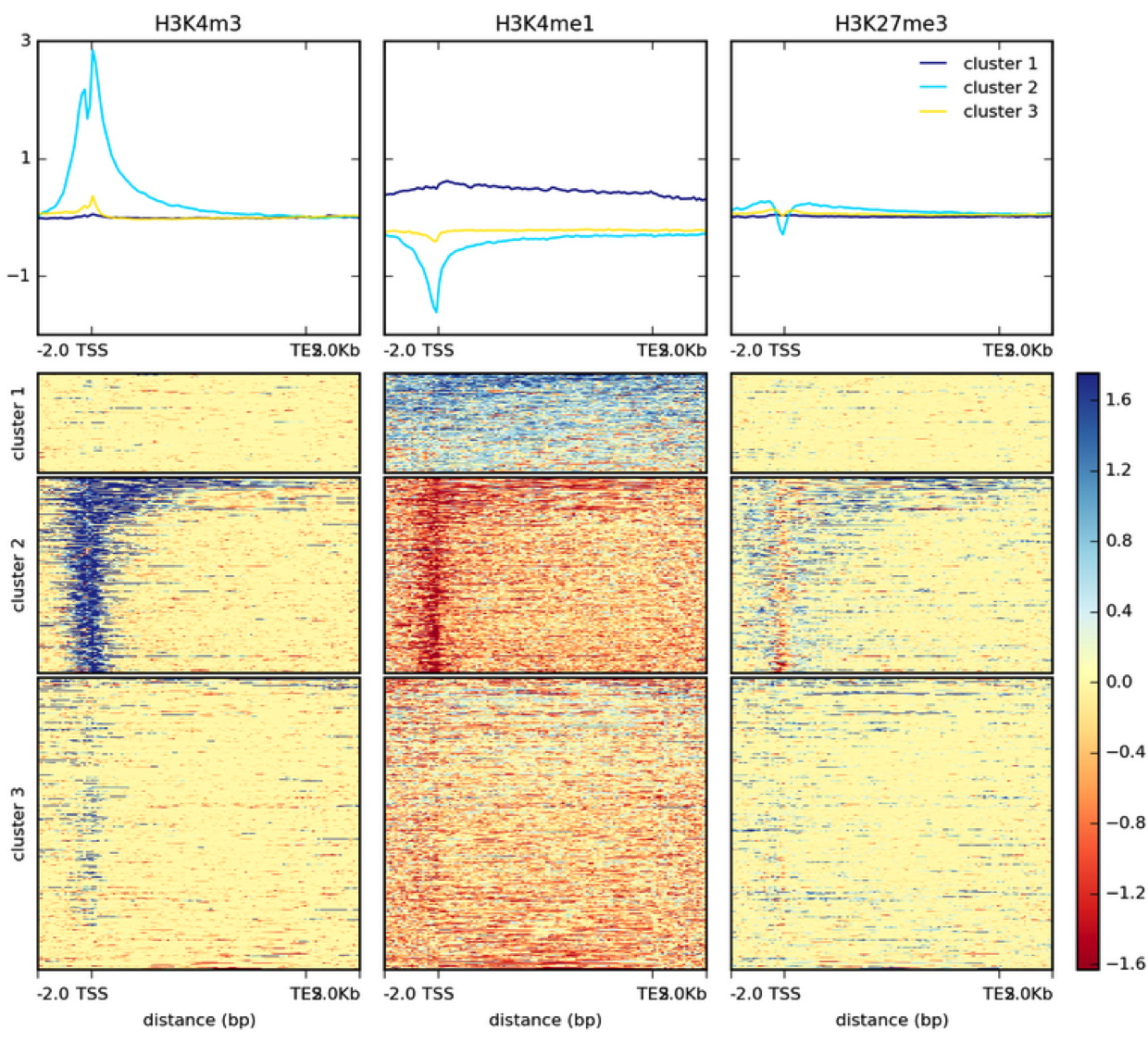
ChIP-seq peak types from various experiments.(a–c) Data shown are from... | Download Scientific Diagram
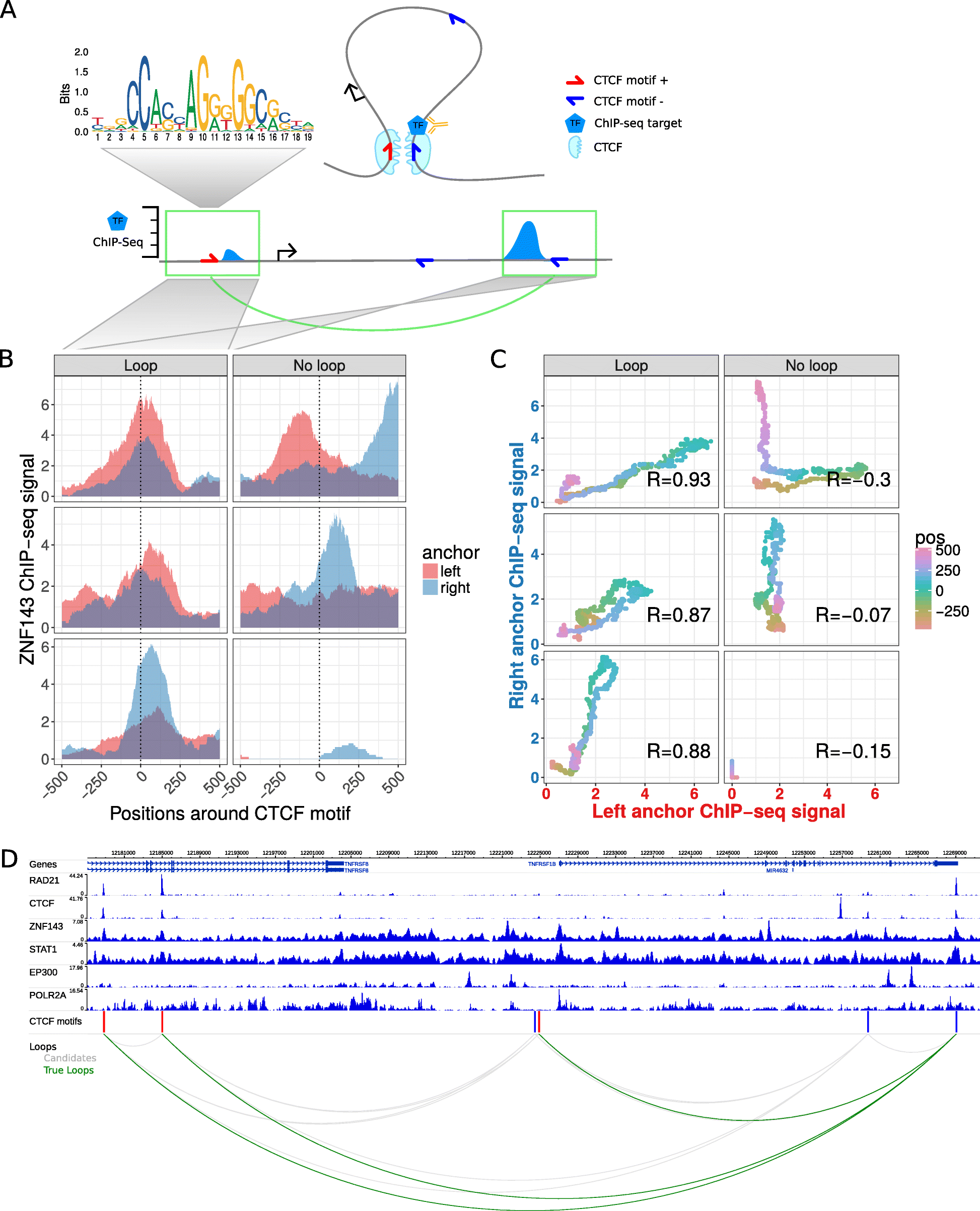
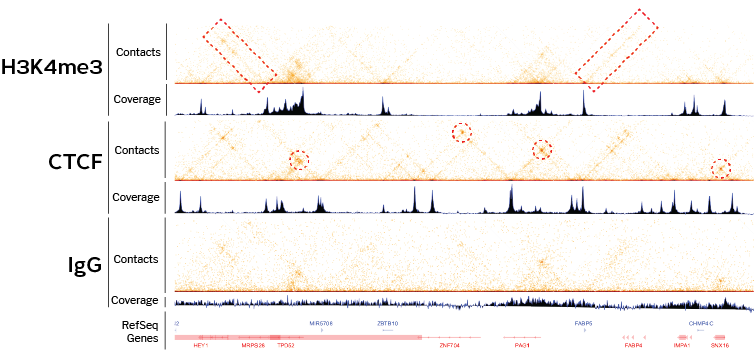
7C: Computational Chromosome Conformation Capture by Correlation of ChIP-seq at CTCF motifs | BMC Genomics | Full Text

Vezf1 protein binding sites genome-wide are associated with pausing of elongating RNA polymerase II | PNAS
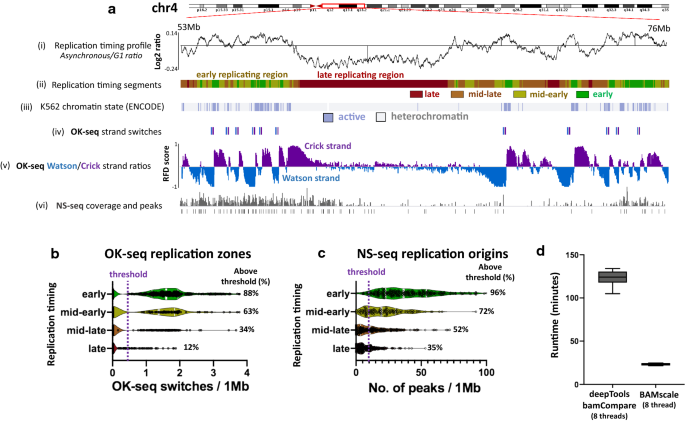
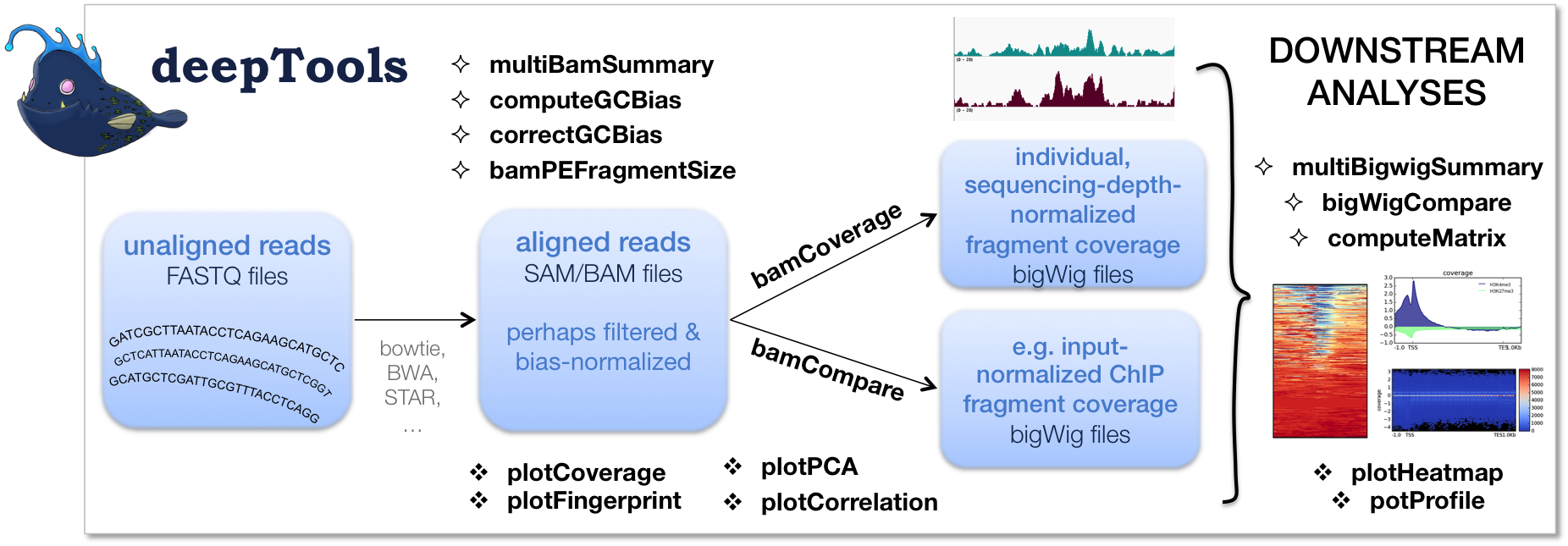
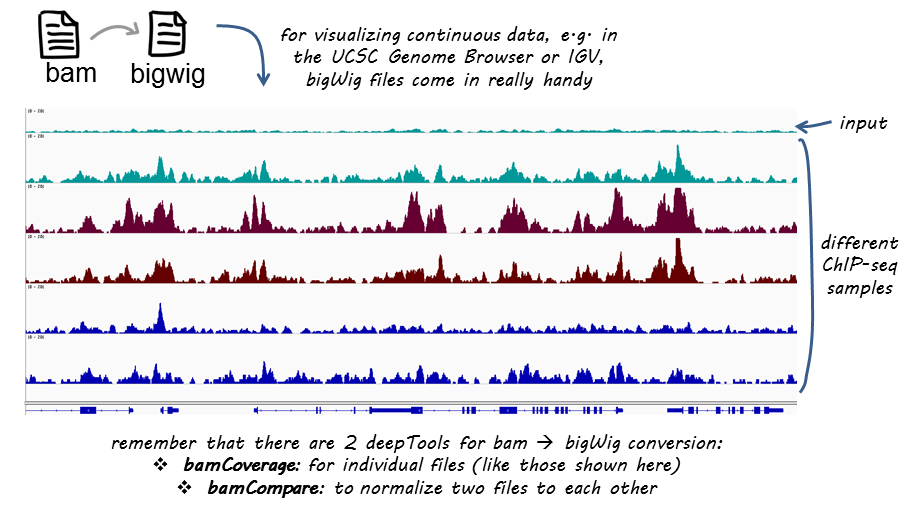
BAMscale: quantification of next-generation sequencing peaks and generation of scaled coverage tracks | Epigenetics & Chromatin | Full Text