![PDF] Cistrome Data Browser: a data portal for ChIP-Seq and chromatin accessibility data in human and mouse | Semantic Scholar PDF] Cistrome Data Browser: a data portal for ChIP-Seq and chromatin accessibility data in human and mouse | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/68bd3de081d1baa8df24bec6ec2ad46df5e48b6c/5-Figure2-1.png)
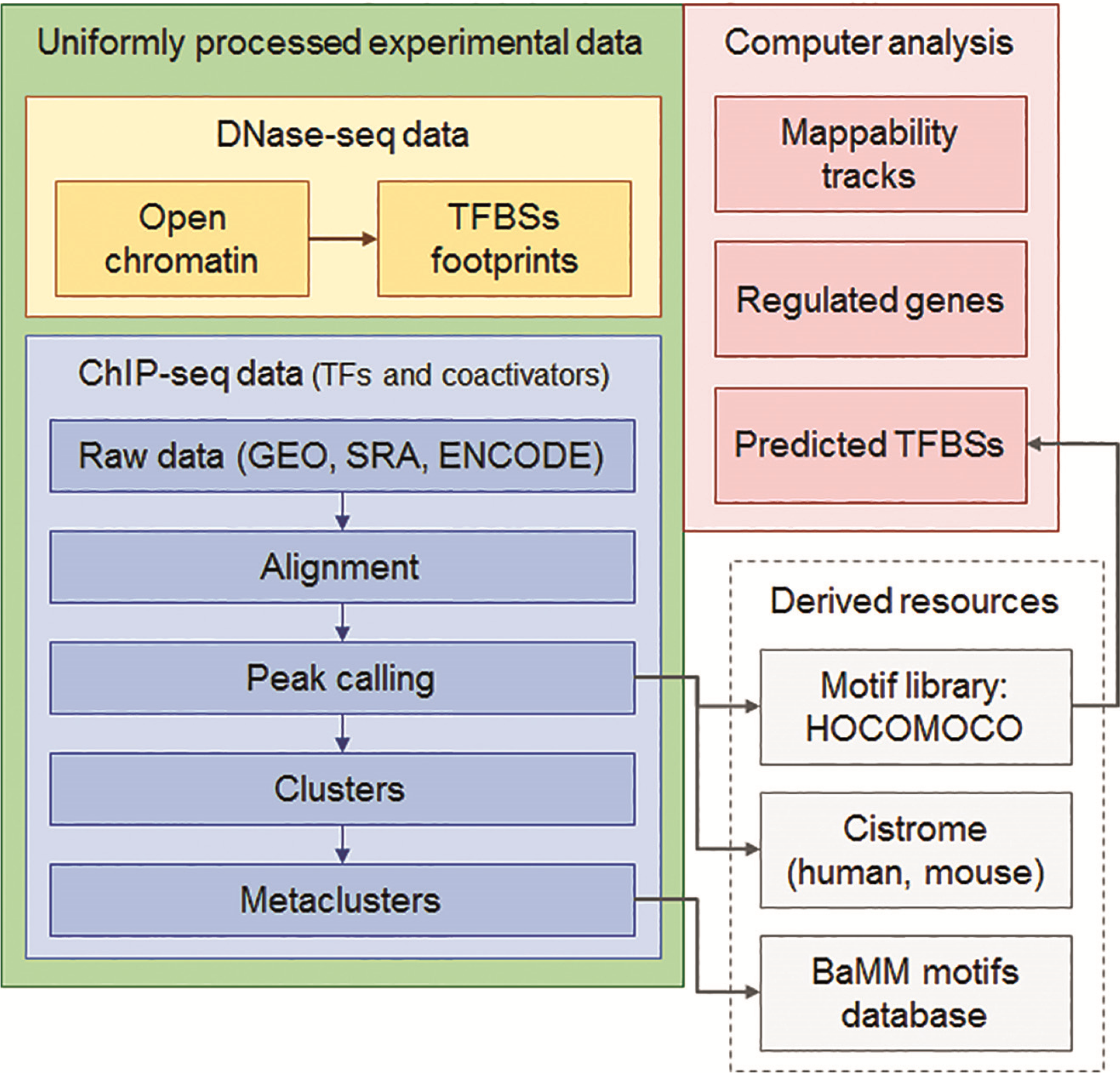
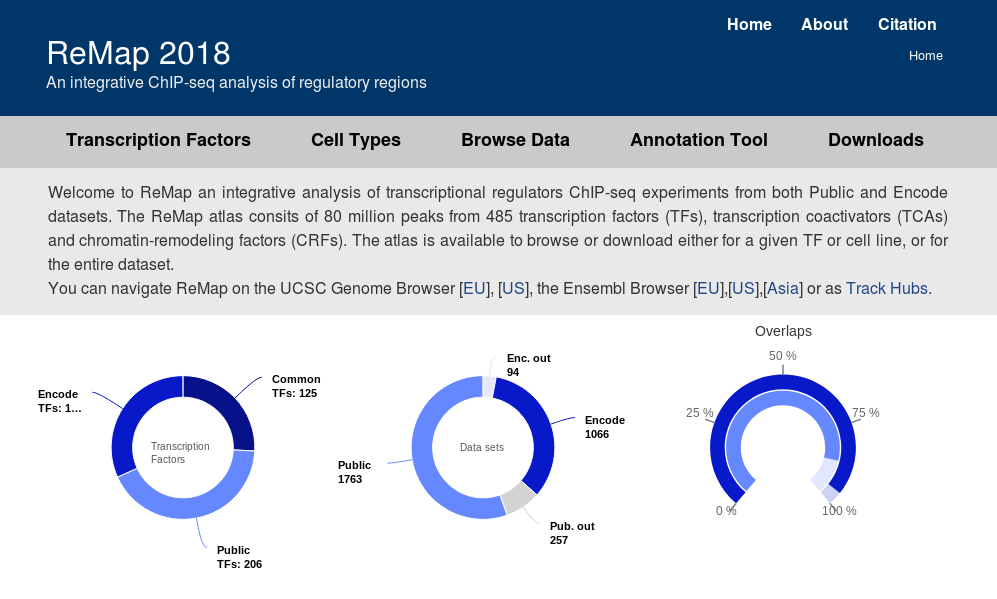
PDF] Cistrome Data Browser: a data portal for ChIP-Seq and chromatin accessibility data in human and mouse | Semantic Scholar

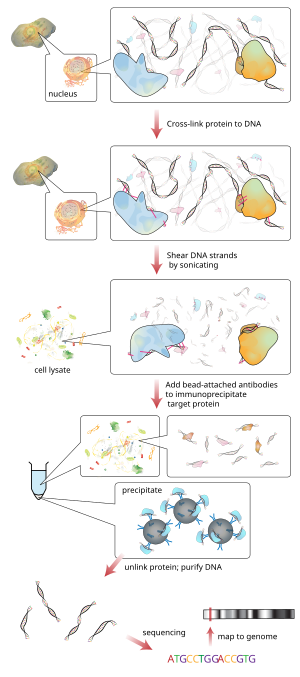
Quantification of Differential Transcription Factor Activity and Multiomics-Based Classification into Activators and Repressors: diffTF - ScienceDirect

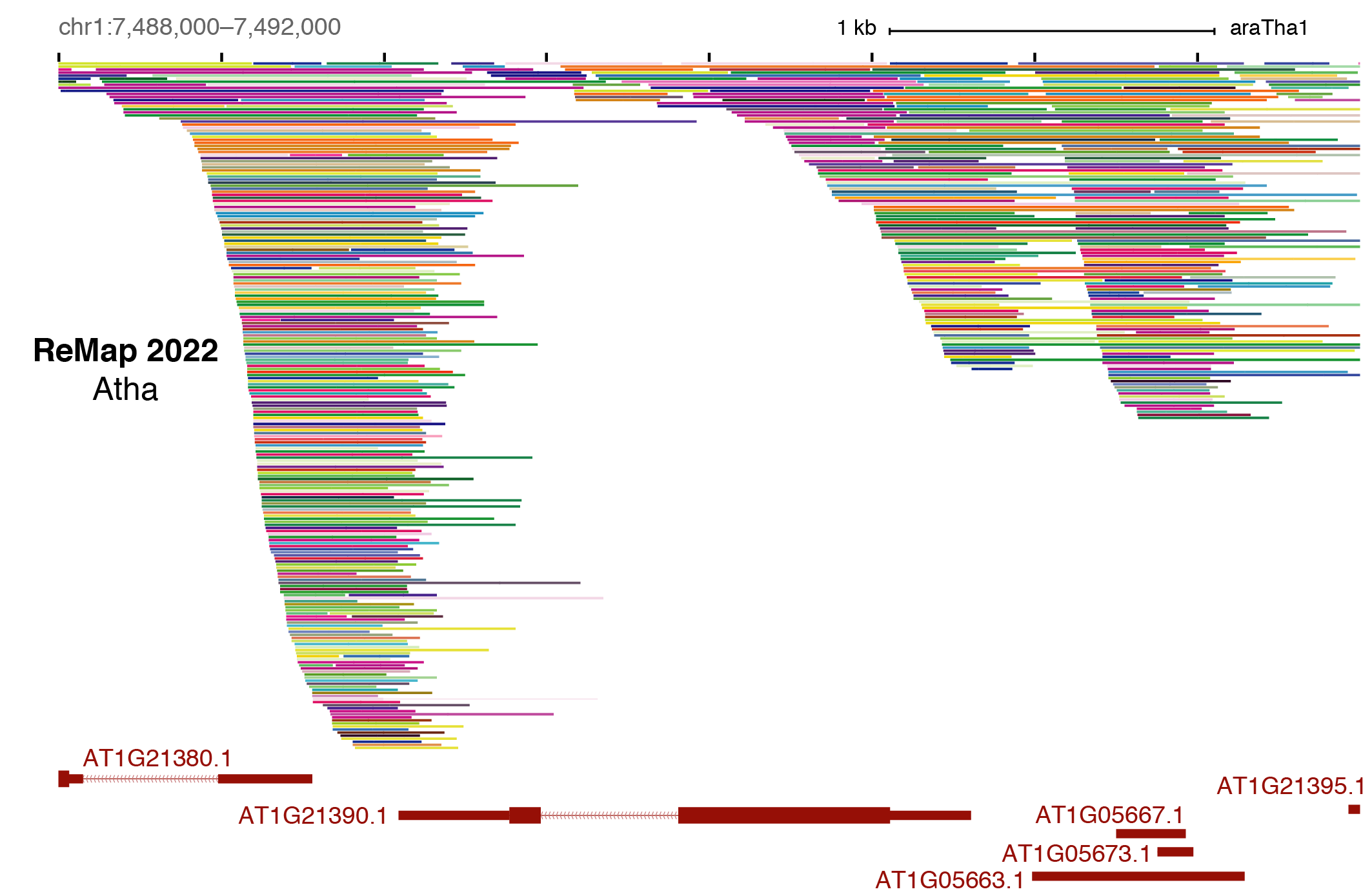
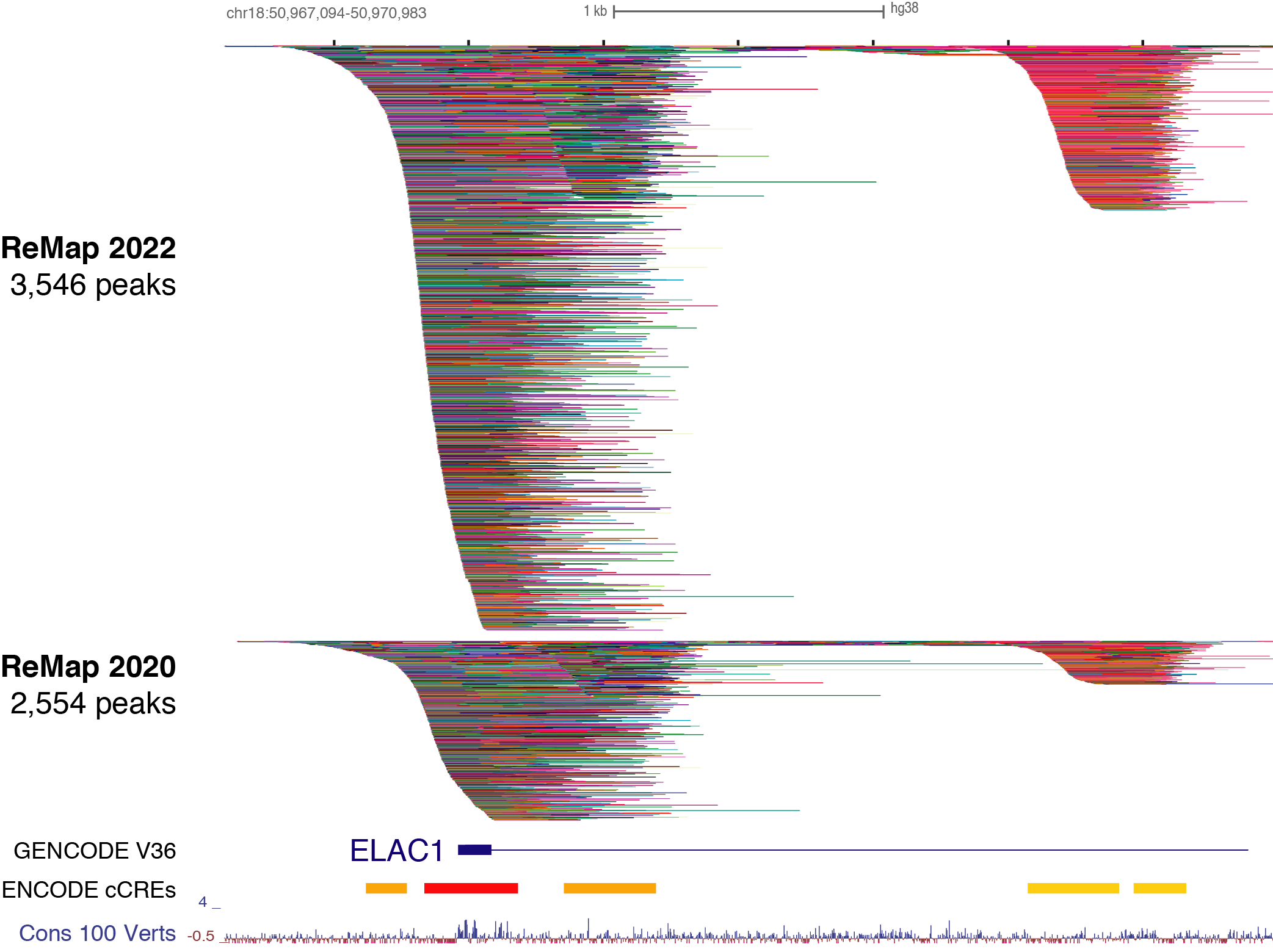
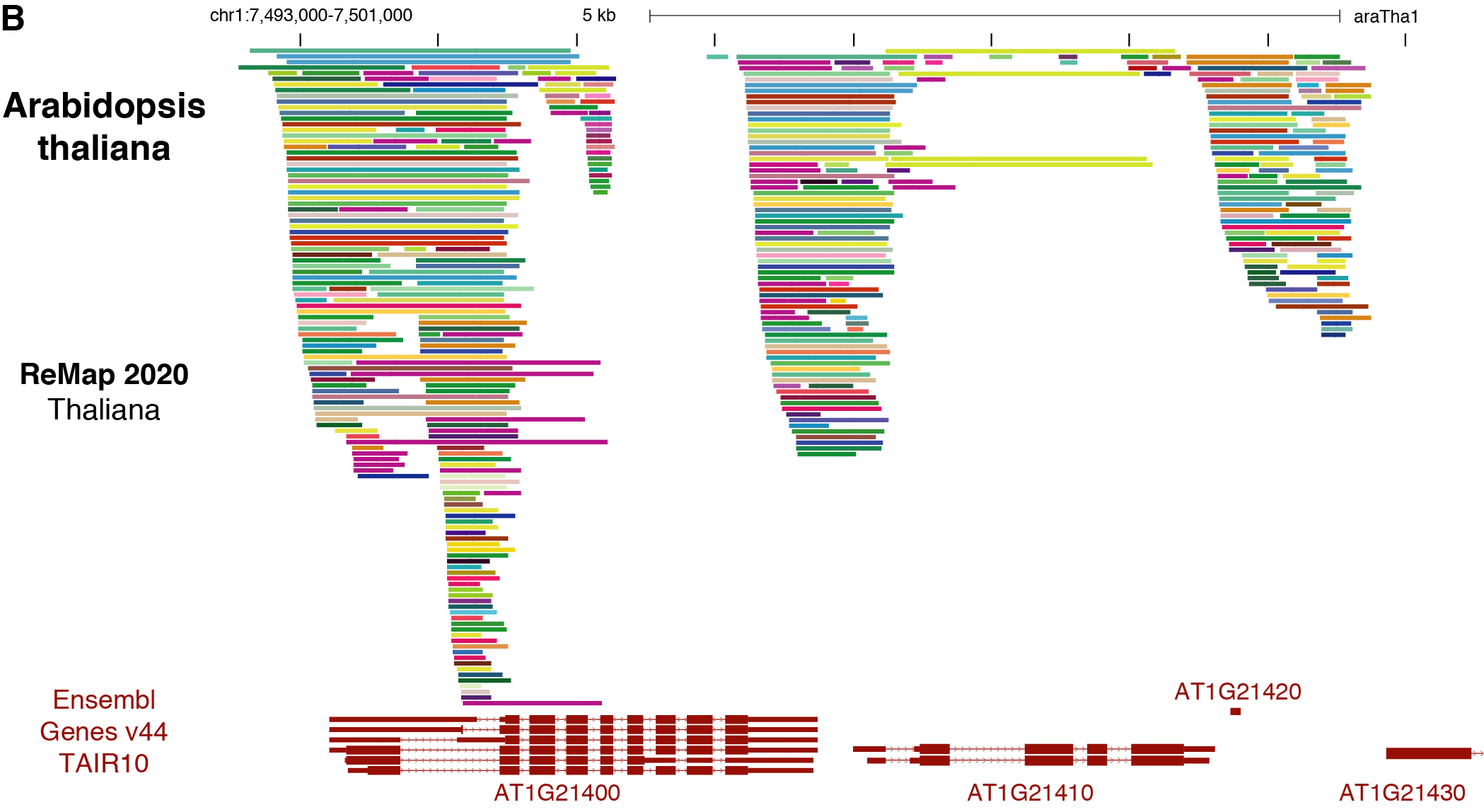
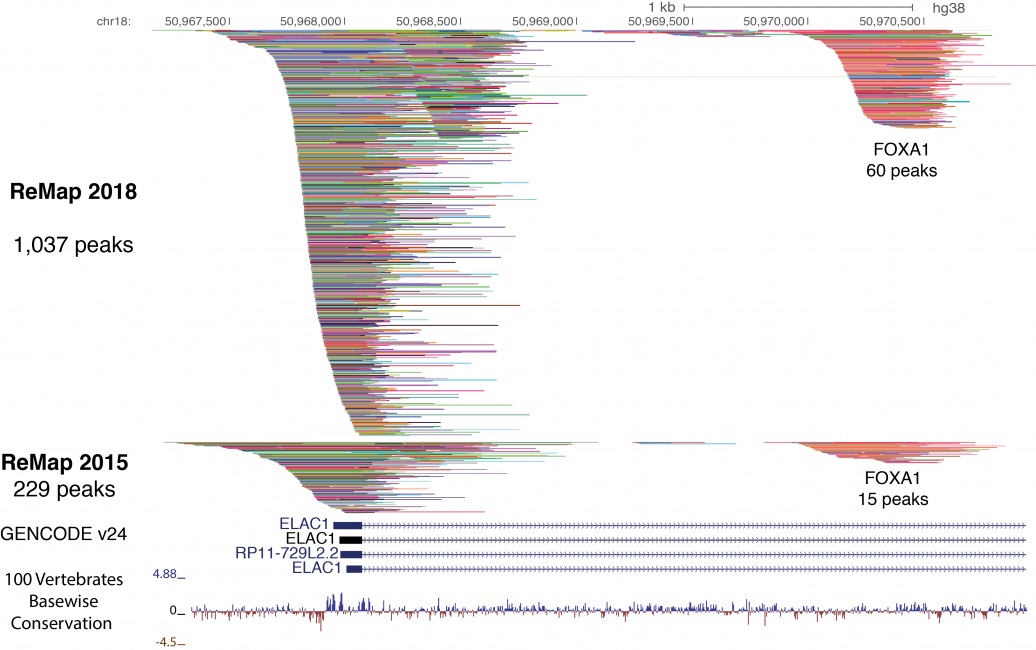
ReMap 2020: a database of regulatory regions from an integrative analysis of Human and Arabidopsis DNA-binding sequencing experiments. - Abstract - Europe PMC

Overview of MANTA2. a) Intersection of the ReMap ChIP-seq regions with... | Download Scientific Diagram
ReMap 2020: a database of regulatory regions from an integrative analysis of Human and Arabidopsis DNA-binding sequencing experi

Mathematics | Free Full-Text | Analysing the Protein-DNA Binding Sites in Arabidopsis thaliana from ChIP-seq Experiments

ChIP-seq peaks and CRMs. (A) A schematic diagram of the three types of... | Download Scientific Diagram
![Predicting stimulation-dependent enhancer-promoter interactions from ChIP- Seq time course data [PeerJ] Predicting stimulation-dependent enhancer-promoter interactions from ChIP- Seq time course data [PeerJ]](https://dfzljdn9uc3pi.cloudfront.net/2017/3742/1/fig-1-full.png)
Predicting stimulation-dependent enhancer-promoter interactions from ChIP- Seq time course data [PeerJ]

ChIP-Seq profiles obtained for ACF1 and RSF-1 in wild-type and mutant... | Download Scientific Diagram
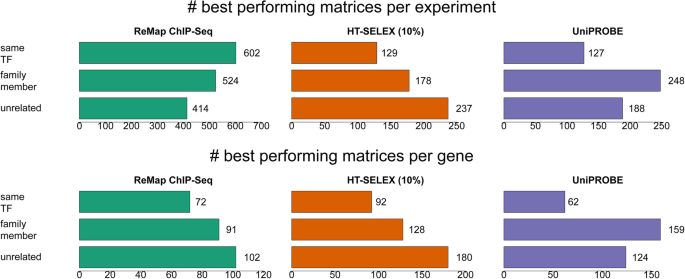
Insights gained from a comprehensive all-against-all transcription factor binding motif benchmarking study | Genome Biology | Full Text
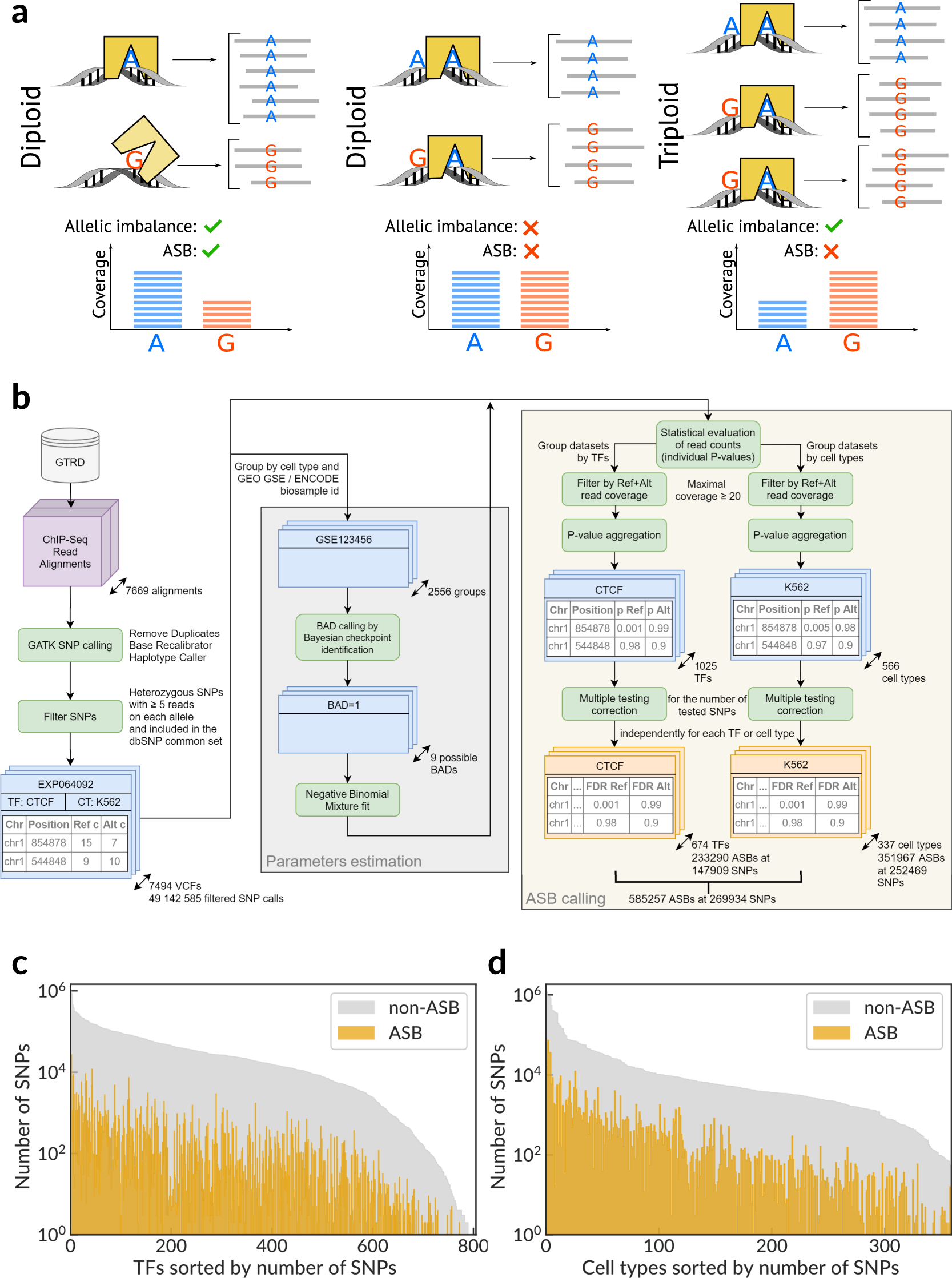
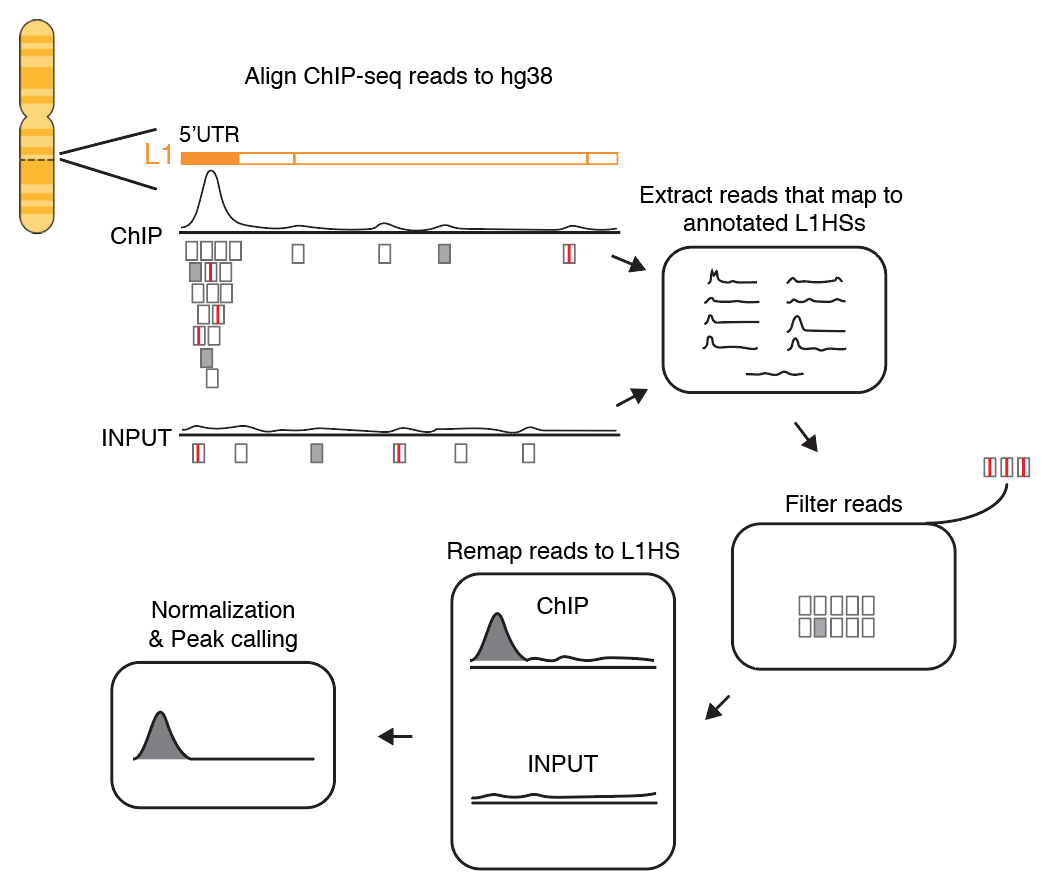
Landscape of allele-specific transcription factor binding in the human genome | Nature Communications